Department of Informatics
University of Bergen
Norway
2. Logical Structure
2.1. Input (HTML page)
2.2. Processing
2.3. Output
Pratt (Jonassen et al, 1995; Jonassen, 1996) is a tool for discovering patterns in sequences. This program takes as input a set of $n$ unaligned sequences, a set of parameters (defining constraints on the class of patterns that can be discovered), and outputs patterns matching some minimum number $k$ ($k$ is chosen by the user) of the $n$ input sequences. The patterns are ranked according to their information content (a measure of the strength of the patterns). Pratt is able to discover patterns of the type used in the PROSITE database. Pratt has earlier been made available to the research community by making the source code (ANSI C) available via anonymous ftp. The motivation for this project was:
The structure of this report is as follows. In Section 2 we describe the logical structure of PrattWWW, and in Section 3 we describe the implementation. The Appendix gives more low-level technical information about the actual programs.
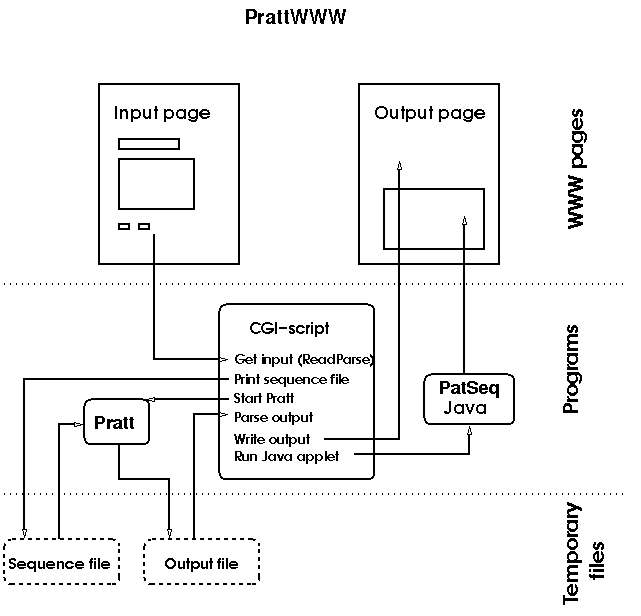
The figure below shows the main components of the PrattWWW system. The WWW pages are shown on the top, and the programs (Pratt, the CGI script, and PatSeq) below, and the information flow is shown with arrows.

The user sets the attributes and parameters with a form. He/she should know a bit about the biological background (Sequences, Patterns), and can choose values for the following parameters:
In the extended version, the user can set all Pratt parameters. (Jonassen et al, 1995; Jonassen, 1996) The possibility is there, but it is not implemented in the processing part yet. For some parameter it is necessary to hand over more values. This must be programmed later.
The user also needs to say which source Pratt should use to get the sequence data. There are 3 possibilities:
The script takes all parameter information from the Web page. If necessary, it stores the pasted sequences in a file. If it is supposed to run Pratt, it constructs a Pratt command line and saves it in a prattcommand file. Afterwards it executes this file. The next step for every alternatives is, to extract the data from the Pratt output file, interprete them and write all necessary information for the presentation in arrays.
The output is splittet in two parts; the textual display (generated by the CGI script programmed in Perl), and the graphical presentation (generated by the Java applet PatSeq).
The first step of the output part is to create and print the head and the title of the output WWW page. The script prepares the structure of the result page
and prints the result of the interpretation to the webbrowser. The general Information are a few sentences to say something about the query. The original Pratt output is stored in a file on the server, it is printed for checking the results. With the query form the user can modify his query or create a new query and start Pratt again.
The CGI script hands over all relevant data about the result to the JAVA applet and calls it. The applet creates with these information an image with 3 parts.
There are some possibilities for interaction between the user and the applet.
When the user
For the tool are used 3 different languages. These are connected; Perl is used to generate HTML code for the output page, and Perl controls the Java applet:
There are two types of HTML WWW pages:
The first type is a HTML page which is stored on the server. This is the Input page (see Section 2.1). It contains a form. With this form, the user can build the query for Pratt as he want, while he sets the parameter for the query. The form refers to the CGI script.
The second type is more complex. After the processing, the browser will show the result at a HTML page. This page is created during the processing. The basic structure of the result pages of different queries is standard (see Section 2.3):
The perl script is structured modular (see Appendix C1). The Main module contains the variables declaration. The script takes the parameter from the Input page (see Section 2.1HTML) to get the values. They are stored in an array for the options and variables for the other parameters. This is realised by the modul 'ReadParse' (created by Steve E. Brenner). The next steps will be executed only if necessary. Dependend on the the input information - where the sequences comes from, the script creates the sequence file and the pratt_command file. They contain respectively the sequence information and the Pratt command line. Then the script executes the pratt_command file.
Afterwards it creates the structure of the output HTML page (see Section 2.3). It calls two of the single parts (detailed result information, new form) submodules. For the presentation of the original it prints the Pratt result file (output.dat) as preformated text.
Now comes the first step in the processing part. It loads the Pratt output file and defines a variable for it. The format of the data file is ASCII. The script splits the data in lines and stores them in an array. Then it executes a 'while' loop for every line. It splits the line in single fields, delimited by space characters and looks for keywords. Each keyword defines a place where the script finds information. The script stores this information in a number of variables and arrays (see Appendix C2).
When this part is finished, the inividual output of the query will be created (see Section 2.3). First the basic information about the query will be printed. Then the script takes the information about the patterns and sequences and stores this information in a set of new arrays (see Appendix C3). The information in these arrays are handed over to the JAVA applet PatSeq.
Next, the script calculates some variables that are important for the dimensioning of arrays in the JAVA applet, and for the physical size of the individual image parts to be generated by the applet (see Appendix C4). Amongst other things, we need to calculate the overlaps of the patterns in each sequence in order to find out how much vertical space is needed for the graphical representation of each sequence. The script calls the JAVA applet with all the necassary data, the parameter transfer is realised with two loops for each sequence and pattern.
In the JAVA applet there are 3 parts of the result to draw.
The code for JAVA applet consists of 3 main parts:
The variables definition should be clear, all necessary variables are be defined. They are organized by the purpose (Pattern, Ruler, Sequence).
In the variables initialisation, the JAVA script gets all parameter which are handed over from the CGI script. The program executes the loops with the running variables (see Appendix D1) to get the data about the different patterns and sequences (see Appendix D2). It is necessary to work with flexible loops, because there can exits different number of Sequences and Patterns in every query. There are also some other necessary variables (see Appendix D3) for the applet. Initially the status of all patterns is set to ON, hence all patterns will be shown in the sequences. The colours (10 different) are initialised using a separe module.
One event handler involves changing the status of individual patterns, and calls a helping module in order to recalculate the coordinates for the drawing of the patterns in the sequences. When new patterns are switched on, new overlaps might result, and consequently patterns may have to be 'pushed down'. Analogously, when patterns are switched on, less vertical space may be needed for each sequence, and hence the view becomes more compact.